Alternative splicing and RNA editing of the sodium channel gene
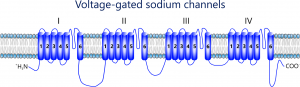
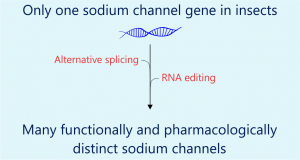
It is well known tha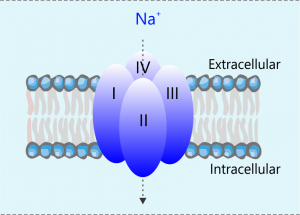 t most mammalian genomes contain ten sodium channel genes, each of which encodes a functional sodium channel of a unique structural, gating, and pharmacological property, as well as a distinct tissue/cell distribution pattern, presumably to fulfill a unique physiological role in specific tissues and cells. In contrast, insect genomes contain only one sodium channel gene. How do insects achieve sodium channel functional diversity? Work from my group and others showed that the functional diversity of sodium channels in insects is achieved by extensive alternative splicing and RNA editing of a single primary transcript.
t most mammalian genomes contain ten sodium channel genes, each of which encodes a functional sodium channel of a unique structural, gating, and pharmacological property, as well as a distinct tissue/cell distribution pattern, presumably to fulfill a unique physiological role in specific tissues and cells. In contrast, insect genomes contain only one sodium channel gene. How do insects achieve sodium channel functional diversity? Work from my group and others showed that the functional diversity of sodium channels in insects is achieved by extensive alternative splicing and RNA editing of a single primary transcript.
 Our work on the splicing and RNA editing of insect sodium channel transcripts set a foundation for us to address several outstanding questions in insect ion channel physiology and neurobiology: How do splicing/RNA editing variants impact neuronal excitability in different cells and tissues? Do these sodium channel variants fulfill different neurophysiological roles in specific neuronal or non-neuronal cells? What would happen to insect development, neurophysiology and behavior if we eliminate certain alternative exons or RNA editing sites in the sodium channel gene? Answering these fundamental questions require precision methods of gene mutations and replacement, which is now feasible using the CRISPR-Cas9 technology.
Our work on the splicing and RNA editing of insect sodium channel transcripts set a foundation for us to address several outstanding questions in insect ion channel physiology and neurobiology: How do splicing/RNA editing variants impact neuronal excitability in different cells and tissues? Do these sodium channel variants fulfill different neurophysiological roles in specific neuronal or non-neuronal cells? What would happen to insect development, neurophysiology and behavior if we eliminate certain alternative exons or RNA editing sites in the sodium channel gene? Answering these fundamental questions require precision methods of gene mutations and replacement, which is now feasible using the CRISPR-Cas9 technology.
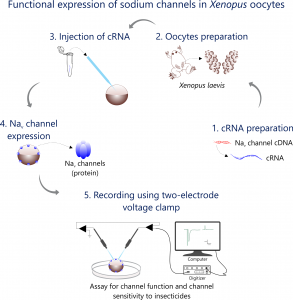
Photos
- Xenopus oocytes preparation, Duke University, February of 2021
- Xenopus oocytes preparation, Duke University, February of 2021